GEMINI: Model Description
GEMINI (Grassland Ecosystem Model with INdividual based Interactions) is a deterministic object-oriented simulation model. It runs in a robust, flexible and portable platform, named UNIF, developed in C++ (C++ Builder 6.0 under Windows for graphical version, and any GCC compliant platform for the command-line version) and is based on a unified numerical integrator. This platform allows the coupling of numerical models. A model is seen as a tree of modules, each having its own numerical integrator variables. These objects are referenced in runtime modifiable lists. A flexible graphical front-end using the VCL library of C++ Builder is generated from the tree of modules.
GEMINI consists of vegetation and soil submodels, coupled with environment and management modules.
Graphical front-end |
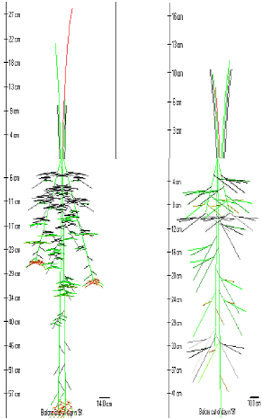
Simulated architecture of F. arundinacea |
The vegetation model, named CANOPT (Soussana and Oliveira-Machado, 2000), is an individual-based model of the growth of mixed pasture species, which describes explicitly the shoot and root morphogenesis of plants and the competition for light and for inorganic N within a multi-layer canopy and soil. It consists of six modules:
- management options (grazing and/or cutting mode, N fertilizer supply);
- environment module, which calculates the microclimate and the inorganic N balance;
- plant growth and allocation module, which simulates the C and N balance and the allocation of growth to the shoot structures, leaf proteins and roots;
- shoot morphogenesis module, which computes the demography and the size of shoots and roots;
- competition module, which calculates radiation (PAR) and N partitioning among plant populations;
- animal module, which calculates the defoliation of each species at grazing and animal returns. The grazing time/bite has 3 components: a fixed prehension time, a mastication time and a time to sort between mixed plant populations.
The C-N soil model (named SOILOPT) describes the dynamics of 4 soil organic compartments, each with a fixed C:N ratio. Two functional microbial groups (e.g. bacteria and fungi) degrade respectively fresh litter and soil organic matter (FOM and SOM decomposers). The 2 microbial groups differ in their potential growth rate, type of substrate used for growth and mineral N requirements. This allows simulation of the energy limitation of SOM decomposers.